Gene Expression: Cervical Cancer
This chapter focuses on two different datasets on cervical cancer, which we will use to perform different tasks. A great in-depth ressource for differential gene expression analysis in R is a mRNAseq workshop by the UC Davis Bioinformatics Core.
Packages
In the upcoming tasks, we will analyse the cervical cancer data from the NBLDA (Negative Binomial Linear Discriminant Analysis) package.
This dataset contains gene expression levels of 714 miRNAs in 29 tumour and 29 non-tumour cervical samples. We aim to determine differences in miRNA gene expression levels between the cervical cancer and non-tumour samples using this dataset.
To perform differential expression analysis of miRNA, we will use limma and edgeR. These are packages distributed via Bioconductor, which is an open source bioinformatics platform.
Installing Packages
# Install BiocManager (Bioconductor's package manager)
if (!require("BiocManager", quietly = TRUE))
install.packages("BiocManager")
# Install limma from Bioconductor
BiocManager::install("limma")
# Install edgeR from Bioconductor
BiocManager::install("edgeR")
# Install NBLDA from CRAN
install.packages("NBLDA")
Loading Packages
# Load the necessary packages
library(NBLDA)
library(edgeR)
library(limma)
Getting Started
To get started, load the cervical dataset into your R session.
# Load the dataset
data(cervical)
Let's take a look at the first 5 rows of the first 3 tumour and non-tumour samples.
> cervical[1:5, c(1:3, 30:32)]
N1 N2 N3 T1 T2 T3
let-7a 865 810 5505 3343 4990 5193
let-7a* 3 12 30 12 99 15
let-7b 975 2790 4912 4217 41617 5152
let-7b* 15 18 27 8 148 14
let-7c 828 1251 2973 519 4664 1274
One can observe, that the tumour/non-tumour information is encoded in the column name "N1" (non-tumour) vs "T1" (tumour).
Data Preprocessing
To analyse the difference in expression, we need to prepare our data beforehand.
Please note that our example is using RNA-seq data.
limma was originally designed for microarray data, but we can also use it for RNA-seq data with its voom() function.
voom() converts RNA-seq data into values that are suitable for linear modelling and differential expression analysis.
This method is also known as the limma voom method.
Choosing the right preprocessing steps according to the data type is crucial in obtaining accurate and reliable results.
If you use data other than RNA-seq, your steps may differ.
1. Creating a DGEList Object
Firstly, we need to create a DGEList (Digital Gene Expression List) object, which is a list that contains various components, including sample information, groups, and genes, among others.
This provides a well-structured and efficient way to manage count data for further analysis.
# Create a DGEList object
cerv0 <- DGEList(cervical)
# Prodide group information
cerv0$samples$group <- substr(colnames(cervical), 1, 1)
2. Data Normalization Factors
We can then calculate the scaling factors with calcNormFactors().
This converts the original library size to a normalized effective library size.
# Compute normalization factors
cerv0 <- calcNormFactors(cerv0)
calcNormFactors() alone does not perform any data normalization.
It concentrates on normalizing the library size.
To achieve complete normalization, you need to include additional steps, such as using the voom() function which we will discuss later.
3. Filtering Low-Expressed Genes
Genes with low expression levels may not contribute significantly to further analysis and may be more affected by noise. We will filter them out in our example.
# see https://ucdavis-bioinformatics-training.github.io/2019_March_UCSF_mRNAseq_Workshop/differential_expression/orig_DE_Analysis.html
# Set the cutoff to "1"
cutoff <- 3
# Calculate the max CPM value for each gene
# Store genes with a max CPM value below the cutoff under "drop"
drop <- which(apply(cpm(cerv0), 1, max) < cutoff)
# Create a new DGEList object by excluding "drop"
cerv <- cerv0[-drop,]
# Look at the originial number of miRNAs
dim(cerv0)
# Look at the number of miRNAs left
dim(cerv)
[1] 714 58
[1] 704 58
We now have expression levels for 704 miRNAs, indicating that 10 miRNAs were removed.
4. Visualizing the Data
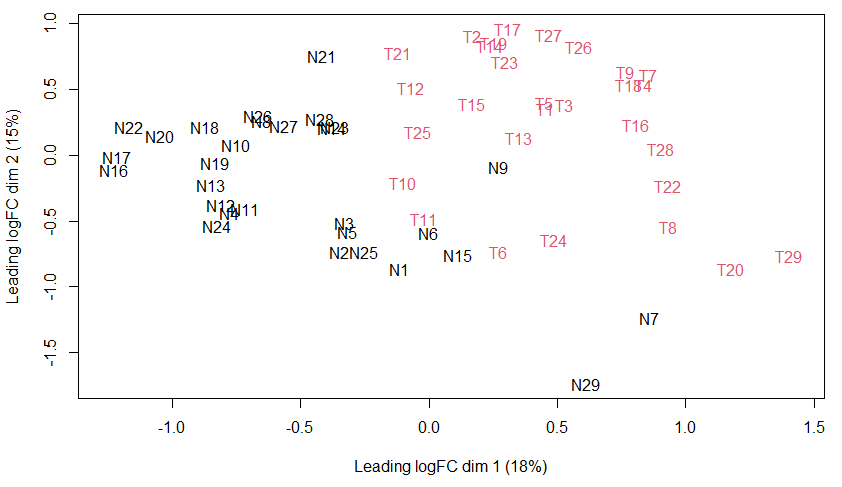
We will go over the most popular approaches for dimensionality reduction: multidimensional scaling (MDS), principal component analysis (PCA), and t-distributed stochastic neighbor embedding (t-SNE). These plots provide a first insight into your gene expression data. Samples that are closer together on the graph have more similar gene expression than those that are further apart. Ideally, samples from the same group should cluster together, indicating that different groups should be separated. Outliers, samples that fall outside the expected range, may indicate poor data quality.
MDS Plot
# Convert the group vector into a numeric vector
# this will convert "N" to 1, "T" to 2
group_numeric <- ifelse(cerv$samples$group == "N", 1, 2)
# Create the MDS plot
# Specify the sample colors based on their groups
plotMDS(cerv, col = group_numeric)
PCA Plot
counts <- t(as.matrix(cerv$counts))
df <- as.data.frame(cbind(cerv$samples$group, counts))
colnames(df)[1] <- "group"
pca_res <- prcomp(counts, scale. = TRUE, center=T)
autoplot(pca_res, colour ="group", data=df) +
theme_classic()+ theme(legend.position="bottom")
tSNE Plot
# Perform tSNE and plot via ggplot. Code adapted from:
# https://stackoverflow.com/questions/44837536/how-to-use-ggplot-to-plot-t-sne-clustering
counts <- t(as.matrix(cerv$counts))
tsne <- Rtsne(counts, perplexity=floor((nrow(counts) - 1) / 3),)
#plot(tsne$Y, col=group_numeric)
tsne_plot <- data.frame(x = tsne$Y[,1], y = tsne$Y[,2],
col = cerv$samples$group)
ggplot(tsne_plot) + geom_point(aes(x=x, y=y, color=col)) +
theme_classic()+ theme(legend.position="bottom")
6. voom-Transformation
Finally, we can perform voom-transformation.
As mentioned above, voom() is a crucial step in the preprocessing of RNA-seq data.
After applying voom(), the data can be used in linear models to perform differential expression analysis.
# Specify the model to be fitted
design <- model.matrix(~ 0 + group)
# voom()
cervoom <- voom(cerv, design, plot = TRUE)
With plot set to TRUE, R will automatically generate a mean-variance plot:
This plot helps you to evaluate if the voom-transformation is appropriate and to check if the variance is stable.
This is necessary for a proper differential expression analysis.
Ideally, the variance should remain constant throughout the range of mean expression values. The dots on the graph should be evenly distributed, forming a horizontal shape or cloud around a straight line. If you notice any noticeable patterns or trends in the graph, this may indicate problems with the normalization or transformation. You should then consider performing additional normalization or transformation steps. If you notice a U- or J-shape, as in our case, this could mean that genes with low expression levels have higher variance. Therefore, it seems that we should filter out more low-expressed genes.
Differential Expression Analysis
After all the preprocessing, we can finally start the differential expression analysis.
1. Fitting Linear Models
Since our data is now appropriate for linear models, we can begin by fitting the model.
# Fit the linear model
fit <- lmFit(cervoom, design)
To perform the differential expression analysis, we then have to create a contrast matrix comparing the non-tumour group to the tumour group.
After that, we need to fit the contrast model.
To improve variances specific to genes, we will use a technique called empirical Bayes moderation (eBayes).
This technique improves the accuracy of differential expression analysis.
# Create a contrast matrix ("N" vs "T")
contrast_matrix <- makeContrasts(groupN - groupT, levels = colnames(coef(fit)))
# Fit the contrast model
fit_contrast <- contrasts.fit(fit, contrast_matrix)
# Apply eBayes()
fit_contrast <- eBayes(fit_contrast)
2. Top-Ranked miRNAs
Lastly, we want to obtain the results of the differential expression analysis.
We can achieve this by using the topTable() function in limma.
# Extract the result
top.table <- topTable(fit_contrast, sort.by = "P", n = Inf)
head(top.table, 10)
This will give you the following table of top-ranked miRNAs:
logFC AveExpr t P.Value adj.P.Val B
miR-7 -3.474128 9.018808 -8.460990 1.262319e-12 5.981448e-10 18.43735
miR-125b 3.676469 13.284352 8.396801 1.680182e-12 5.981448e-10 18.15492
miR-10b* 3.900269 6.003784 8.186562 4.284794e-12 1.016924e-09 17.24763
miR-140-5p 2.606562 9.308576 8.073335 7.091159e-12 1.220002e-09 16.76360
miR-143 3.497444 15.738198 8.030807 8.567429e-12 1.220002e-09 16.57698
miR-205 -5.685219 9.916617 -7.764754 2.792235e-11 3.313452e-09 15.43504
Candidate-12-3p -3.196413 5.167992 -7.702141 3.685426e-11 3.748605e-09 15.16657
miR-944 -3.961094 5.439555 -7.665702 4.331013e-11 3.854602e-09 15.01042
miR-125a-5p 2.961733 10.762754 7.638651 4.882114e-11 3.862295e-09 14.89378
miR-21* -3.675837 8.285200 -7.565493 6.748198e-11 4.560699e-09 14.57877
topTable() ColumnslogFC: The logarithm of the fold change indicates the difference in gene expression between the tumour and non-tumour group. Positive values show upregulation in tumour samples, while negative values show downregulation in tumour samples.AveExpr: The average gene expression level across all samples.t: The t-statistic measures the difference in the means of gene expression between tumour and non-tumour samples, which has been normalized by the standard error.P.Value: The raw p-value.adj.P.Value: The adjusted p-value.B: The B-statistic is associated with the posterior probability that a gene is expressed differentially. Positive B-values provide evidence for differential expression.
We have now identified ten potentially differentially expressed miRNAs between non-tumour and cervical cancer samples. "Potentially" indicates that further analysis is required to confirm these findings. It is always important to interpret bioinformatic analyses with caution.
Sources & Further Reading
- https://ucdavis-bioinformatics-training.github.io/2019_March_UCSF_mRNAseq_Workshop/differential_expression/orig_DE_Analysis.html
- Cerami et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discovery. May 2012 2; 401.
- Cervical Cancer (MSK, 2023). Targeted Sequencing of 177 cervical tumours and their matched normal samples via MSK-IMPACT. https://www.cbioportal.org/study/clinicalData?id=cervix_msk_2023.
- Chen Y, Lun ATL, Smyth GK (2016). From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Research 5, 1438
- Gao et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013).
- Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S. (2005). Bioinformatics and Computational Biology Solutions Using R and Bioconductor. doi.org/10.1007/0-387-29362-0
- Goksuluk D, Zararsiz G, Korkmaz S (2022). NBLDA: Negative Binomial Linear Discriminant Analysis. R package version 1.0.1, https://CRAN.R-project.org/package=NBLDA.
- Kassambara A, Kosinski M, Biecek P (2021). survminer: Drawing Survival Curves using 'ggplot2'. R package version 0.4.9, https://CRAN.R-project.org/package=survminer.
- Law CW, Chen Y, Shi W, Smyth GK (2014). Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biology 15, R29. doi:10.1186/gb-2014-15-2-r29.
- McCarthy DJ, Chen Y and Smyth GK (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Research 40, 4288-4297
- Morgan M, Ramos M (2023). BiocManager: Access the Bioconductor Project Package Repository. R package version 1.30.21, https://CRAN.R-project.org/package=BiocManager.
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015). “limma powers differential expression analyses for RNA-sequencing and microarray studies.” Nucleic Acids Research, 43(7), e47. doi:10.1093/nar/gkv007.
- Robinson MD, McCarthy DJ and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139-140
- Therneau T (2023). A Package for Survival Analysis in R. R package version 3.5-5, https://CRAN.R-project.org/package=survival.
- Witten D, et al. (2010) Ultra-high throughput sequencing-based small RNA discovery and discrete statistical biomarker analysis in a collection of cervical tumours and matched controls. BMC Biology, 8:58. Published online 2010 May 11. doi: 10.1186/1741-7007-8-58